Continuing a common theme of this blog – free or cheap software for scientific purposes, I thought I’d give a quick run-down of the free software available for segmenting CT data into digital models (e.g. objs/stls). This list is by no means comprehensive, but I’ve tried everything here to a reasonable degree. I’m not planning on doing tutorials or anything for these (I did make a youtube video/blog post for using 3D slicer) unless there’s significant demand. Instead, I have tried everything below (up to “other shoutouts”) with a crocodile hindlimb dataset; I’ll give you a couple of screenshots of the interface, and tell you how easy I found them to use.
3D Slicer
- Link: https://www.slicer.org/
- Version tested: 4.10.1
- Pros: Pretty powerful, logical interface, seems to be the most commonly used free CT software on PC.
- Cons: Probably way more powerful than any one person needs, and this can be a bit daunting. Slows down quite a bit with micro-CT data.
- Interface:

I’ve been using 3D Slicer on and off since I was doing my PhD over a decade ago. To be honest, it doesn’t really feel like the software has changed that much in that time, but I think a lot of the development has gone into plugins and performance, which makes sense; version 3 was released in 2007, and we’re only on version 4.10 now in 2019.
That’s not to say it’s old and clunky – far from it, it’s one of the nicer interfaces to use today. It handles image stacks in tiff, jpg, dicom etc, and generally runs pretty quick. I have had issues with micro-Ct tiff stacks in 3DSlicer, but that goes for most of the software here unless you downsample/crop them before processing. Be sure to check out my tutorial here, and this video, which takes you through to 3D printing:
MeVisLab
- Link: https://www.mevislab.de/
- Version tested: 3.11
- Pros: Super powerful node-based workflow. Stunning renderer built in for amazing stills and animations directly from the software.
- Cons: Documentation is not great, and there are few examples of useful [to me] pipelines online. Basically, MeVisLab has one hell of a steep learning curve.
- Interface:

I discovered MeVisLab a few years ago, and have kept an eye on it since. It’s not easy to use, but the node-based workflow will be familiar to people that have used Avizo/Amira or AVS. I think there’s a lot of power in there, but I do struggle to really make the most of it. Mainly, I’ll pull out MeVisLab when I want to make some pretty images, because it’s CUDA-based pathtracer is phenomenal:

And here’s a better one from a different dataset (that I can’t replicate at the moment, because the workflow is so complicated!):
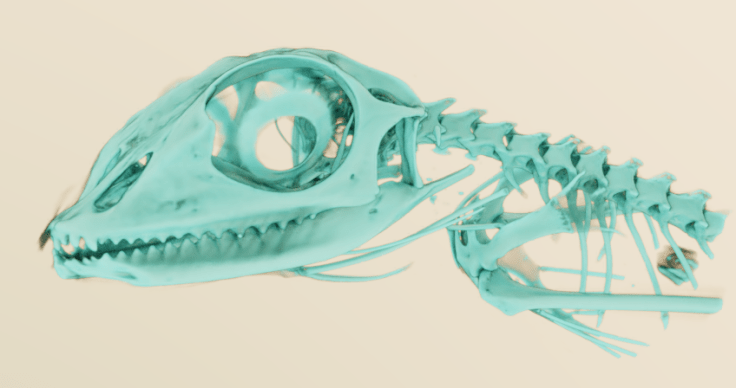
Seg3D 2
- Link: http://www.sci.utah.edu/cibc-software/seg3d.html
- Version tested: 2.4.4
- Pros: Simple to use, Otsu thresholding tends to work really well to get you bones and soft-tissues in just a couple of button clicks.
- Cons: Export doesn’t smooth the mesh at all, so you end up with a blocky mesh (each block being a voxel). This also takes a long time to export. So it’s more useful for exporting masks into another piece of software than it is making models directly
- Interface:

Seg3D 2 has, like a few of the other options here, a nice clean interface (dark mode!). Volume rendering is pretty snappy, and as mentioned in the pros above, Otsu thresholding can get you bones and soft tissue as separate models very quickly. Tools for pulling out individual bones are limited, in my experience.
Dragonfly
- Link: http://www.theobjects.com/dragonfly/
- Version tested: 4.0
- Pros: Pretty good interface, logical to use. 3D volume view is very fast. “Deep learning” stuff.
- Cons: doesn’t seem quite as flexible as 3D Slicer, yet to find a way to easily separate bones. Free for non-commercial academic use only.
- Interface:

Dragonfly is the newest of the software packages I’ve tried. I really like the interface, but appreciate it might be information overload for some. Dragonfly has some awesome deep learning stuff that I don’t really understand, but I do know you can segment all the bones, close 3D areas (to capture insides of bones) then use an algorithm to segment out cortical and trabecular bone separately. Which might be useful to some. If you segment all the bones then run an ROI analysis, you can get it to separate out all the islands, resulting in mostly individual bones. It can export nice 3D models in a range of formats. It’s pretty good!
InVesalius
- Link: https://invesalius.github.io/
- Version tested: 3.1.1
- Pros: Easy to use, clean interface
- Cons: Little bit slower than some of the others in terms of loading and visualizing data.
- Interface:
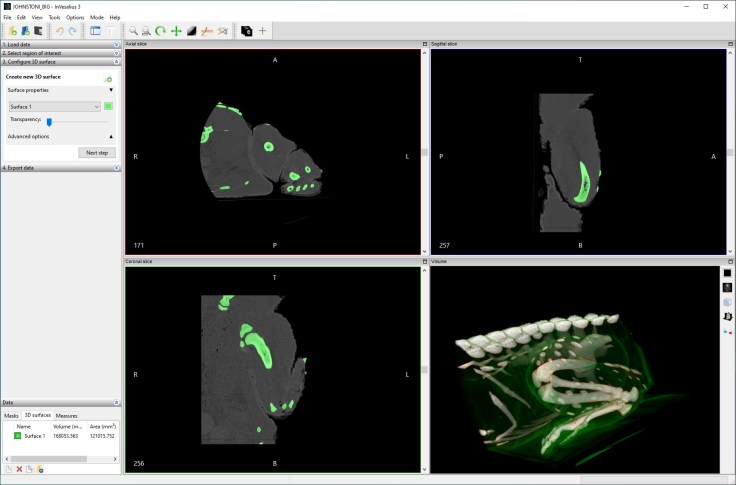
It’s a nice clean interface with some basic thresholding available from the get-go. Manual painting, along with hole filling (automatic and manual) are also available.
There’s also a ‘select part’ tool (and ‘remove part’) which lets you pick individual bones based on connected voxels. I could really quickly pull out individual bones in the leg, for instance.
Exported model (of the bones) was of really high quality. Really nice and easy to use software to export models quickly.
Other shoutouts:
Horus Project
- Link: https://horosproject.org/
- Pros: Looks great. Practically everyone with a Mac uses this.
- Cons: Mac only, and the philosophy of the developers is to try and get you to pay for virtual machines if you don’t have a mac.
- Interface: Can’t use it on my computer, so go look at the website!
I have nothing to say here because I’ve never used it, being firmly in the PC camp. It’s just about the only piece of software that’s made me consider a mac (or at least hackintosh) because when I see people using it, it looks great. But I don’t have a mac, so I can’t tell you any more about it!
Spiers.
Developed by Palaeontologists Mark Sutton, Russell Garwood, and Alan Spencer, Spiers was originally developed for reconstructing 3D fossils from serial sections. The process works for CT data too. It’s actually a package of several applications: View, Edit, and Align. Hopefully your CT data is already aligned! I couldn’t test it with this dataset, because it currently doesn’t import DICOM data. I’ve played with it before but I don’t think it’s designed with quick thresholding and exporting to models in mind (though I could be wrong there).
ImageJ
ImageJ can be used for basic thresholding and volume rendering from image stacks. I don’t use it much though, so can’t tell you much more.
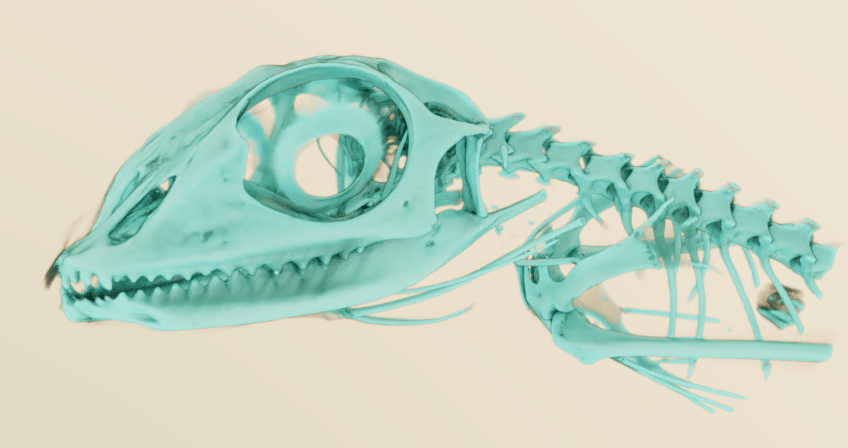
I dearly want to try out MeVisLab but even with some background in Amira, I just can’t figure it out…
Yeah, it’s pretty impenetrable. I’ll write up a post with some example networks at some point, though these days I’ve turnd to Dragonfly for my CT segmentation needs.
Can I ask, is it even possible to load a TIFF stack in MeVisLab to produce vol visualisations, or do the files need to be converted into another format? So far I can only find how to import each TIFF image individually into the workspace.
Yes, you absolutely can load TIFF stacks (in fact it’s tiff handling was why I started using it and foisted it on my PhD student)!
Unintuitively, you use DirectDicomImport to import the TIFF stack (import non-DICOM in the settings for that).
Take a look at these:
Is the quality of the meshes produced by dragonfly any good?
Yeah, its great. Probably as good or better than anything else I’ve used.
3D slicer is great and it does so much more than just segmentation. We use it regularly. But for Deep Learning to aid segmentation, take a look at RILcontour.
I am working on AI in the browser for segmentation with tensorflowjs. Here is a super early prototype: http://www.medicalsegmentation.com.
Thanks, I’ll take a look at those
Thank you so much this is one of the best recommendation lists i have seen and i look forward to trying new options — you made me happy today – thank you